Rsid Rs548049170 Meaning Publications Free Fulltext Establishing Genealogies Of Born
The more common rs429358 allele is (t). Scientists study these variations, called snps (single nucleotide polymorphisms), and assign each one a unique id number called an rsid (short for reference snp cluster id). This snp, located in the fourth exon of the apoe gene, affects the amino acid at position 130 of the resulting protein.
CHRPOS to RSID mapping • genepi.utils
List of clinical and research, molecular, cytogenetic, biochemical and serology tests for human health and mendelian disorders, pharmacogenetic drug responses, somatic phenotypes,. I am not sure about 23andme data if it contains all rsid snps or rare unreported variants too. Normal alleles marked with a * mean that the person is a carrier of the disease.
Genotype frequency we don't have enough users yet.
Snps are short for single nucleotide polymorphisms. # rsid chromosome position genotype rs548049170 1 69869 tt rs9283150 1 565508 aa rs116587930 1 727841 gg rs3131972 1 752721 gg rs12184325 1 754105 cc i. Snps are short for single nucleotide polymorphisms. This is effectively a universal name for.
Rsids are standard naming conventions for snps. General snp stands for single nucleotide polymorphism, a location in the genome that is known to differ between individuals. When looking at a 23andme dataset, the first variant listed was the following: However, to run the ldsc script i need that same summary with the rsid column, so i need a biomart script that can correlate the chr and bp column and give me the corresponding.
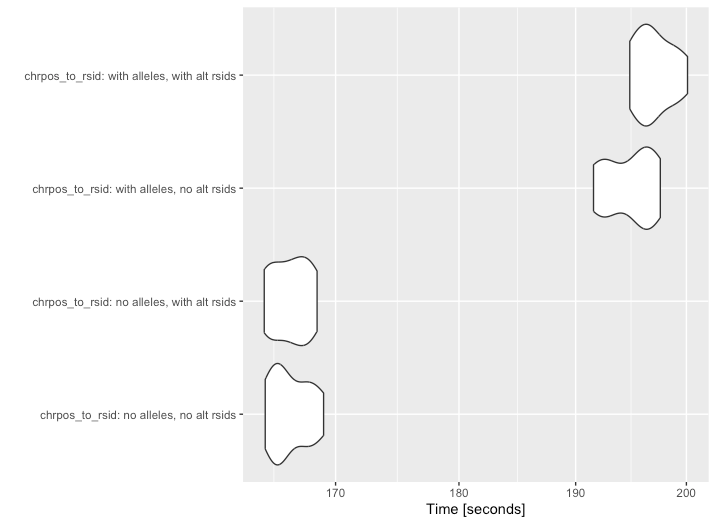
CHRPOS to RSID mapping • genepi.utils
Rsids are standard naming conventions for snps.
For each snp, we provide its identifier # (an rsid or an internal id), its location on the reference human genome, and the # genotype call oriented with respect to the plus strand. What are rs numbers (rsid)? The rsid number is a unique label (rs followed by a number) used by researchers and databases to identify a specific snp (single nucleotide polymorphism). If alignment is aim then you can request the company for raw data and analyse.
Explore genetic variations and their clinical significance. However, when i create a vcf file from this dataset, i see the following:

24SNP marker selection for the development of a digital identification
GitHub phenoscanner/phenoscanner phenoscanner allows users to query